-
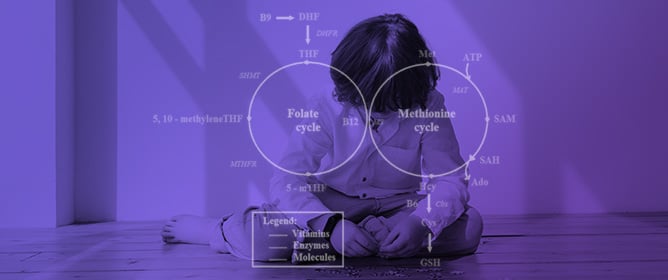 Folate–Methionine Cycle Disruptions in ASD Patients and Possible Interventions: A Systematic Review
Folate–Methionine Cycle Disruptions in ASD Patients and Possible Interventions: A Systematic Review -
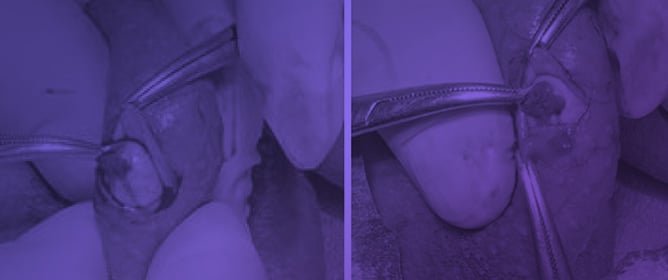 The Klinefelter Syndrome and Testicular Sperm Retrieval Outcomes
The Klinefelter Syndrome and Testicular Sperm Retrieval Outcomes -
 Historical Mitogenomic Diversity and Population Structuring of Southern Hemisphere Fin Whales
Historical Mitogenomic Diversity and Population Structuring of Southern Hemisphere Fin Whales -
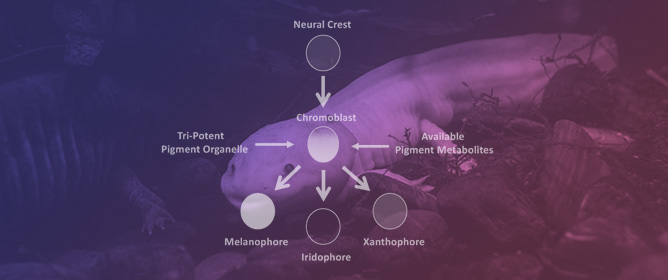 Leukocyte Tyrosine Kinase (Ltk) Is the Mendelian Determinant of the Axolotl Melanoid Color Variant
Leukocyte Tyrosine Kinase (Ltk) Is the Mendelian Determinant of the Axolotl Melanoid Color Variant -
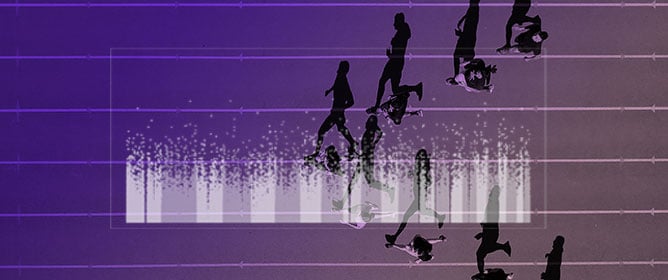 Exome-Wide Association Study of Competitive Performance in Elite Athletes
Exome-Wide Association Study of Competitive Performance in Elite Athletes
Journal Description
Genes
Genes
is a peer-reviewed, open access journal of genetics and genomics published monthly online by MDPI. The Spanish Society for Biochemistry and Molecular Biology (SEBBM) is affiliated with Genes and their members receive discounts on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Genetics & Heredity) / CiteScore - Q2 (Genetics)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 17.9 days after submission; acceptance to publication is undertaken in 2.6 days (median values for papers published in this journal in the first half of 2023).
- Recognition of Reviewers: Reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
3.5 (2022);
5-Year Impact Factor:
3.9 (2022)
Latest Articles
The Genetic Diversity and Genetic Structure of the Germplasm Resources of the Medicinal Orchid Plant Habenaria dentata
Genes 2023, 14(9), 1749; https://doi.org/10.3390/genes14091749 (registering DOI) - 01 Sep 2023
Abstract
Habenaria dentata has medicinal and ornamental value, but the number of wild populations is decreasing dramatically. Thus, conducting research on its genetic diversity and structure is necessary to provide a basis for its conservation. This study aimed to explore the genetic diversity of
[...] Read more.
Habenaria dentata has medicinal and ornamental value, but the number of wild populations is decreasing dramatically. Thus, conducting research on its genetic diversity and structure is necessary to provide a basis for its conservation. This study aimed to explore the genetic diversity of the wild plant H. dentata and protect and optimize its wild resources. The genetic diversity of 133 samples from six wild populations of H. dentata was analyzed using Inter Simple Sequence Repeat molecular markers to provide a scientific basis for the screening of improved germplasm resources. The results showed that the average number of alleles was 1.765, the average number of effective alleles was 1.424, the average Nei’s gene diversity index was 0.252, the average Shannon diversity index was 0.381, and the average percentage of polymorphic loci was 76.499%. The variation within the populations was 77.34%, and the variation between the populations was 22.66%. The gene flow was 1.705, which was greater than 1. The results of the cluster analysis showed that the six populations were mainly divided into four clusters and were not classified according to their geographical location. There was no significant correlation between the geographical location and genetic distance between the populations (r = 0.557, p > 0.05). The genetic diversity of H. dentata is high. Among the six wild populations, the genetic diversity of the Mulun population was the highest and this population can be used as a key protection unit. The study on the genetic diversity of H. dentata can not only reveal the reasons for the decrease in the number of individuals in the population to a certain extent, and put forward the protection strategy, but also provide a scientific basis for the breeding of excellent seed resources.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►
Show Figures
Open AccessFeature PaperArticle
The Rosetta Phenotype Harmonization Method Facilitates Finding a Relationship Quantitative Trait Locus for a Complex Cognitive Trait
by
, , , , , , , and
Genes 2023, 14(9), 1748; https://doi.org/10.3390/genes14091748 (registering DOI) - 31 Aug 2023
Abstract
Genetics researchers increasingly combine data across many sources to increase power and to conduct analyses that cross multiple individual studies. However, there is often a lack of alignment on outcome measures when the same constructs are examined across studies. This inhibits comparison across
[...] Read more.
Genetics researchers increasingly combine data across many sources to increase power and to conduct analyses that cross multiple individual studies. However, there is often a lack of alignment on outcome measures when the same constructs are examined across studies. This inhibits comparison across individual studies and may impact the findings from meta-analysis. Using a well-characterized genotypic (brain-derived neurotrophic factor: BDNF) and phenotypic constructs (working memory and reading comprehension), we employ an approach called Rosetta, which allows for the simultaneous examination of primary studies that employ related but incompletely overlapping data. We examined four studies of BDNF, working memory, and reading comprehension with a combined sample size of 1711 participants. Although the correlation between working memory and reading comprehension over all participants was high, as expected (
(This article belongs to the Section Human Genomics and Genetic Diseases)
Open AccessEditorial
The Stability and Evolution of Genes and Genomes
Genes 2023, 14(9), 1747; https://doi.org/10.3390/genes14091747 - 31 Aug 2023
Abstract
The existence of current species can be attributed to a dynamic interplay between evolutionary forces and the maintenance of genetic information [...]
Full article
(This article belongs to the Special Issue The Stability and Evolution of Genes and Genomes)
Open AccessArticle
A Homozygous MAN2B1 Missense Mutation in a Doberman Pinscher Dog with Neurodegeneration, Cytoplasmic Vacuoles, Autofluorescent Storage Granules, and an α-Mannosidase Deficiency
by
, , , , , , , and
Genes 2023, 14(9), 1746; https://doi.org/10.3390/genes14091746 - 31 Aug 2023
Abstract
A 7-month-old Doberman Pinscher dog presented with progressive neurological signs and brain atrophy suggestive of a hereditary neurodegenerative disorder. The dog was euthanized due to the progression of disease signs. Microscopic examination of tissues collected at the time of euthanasia revealed massive accumulations
[...] Read more.
A 7-month-old Doberman Pinscher dog presented with progressive neurological signs and brain atrophy suggestive of a hereditary neurodegenerative disorder. The dog was euthanized due to the progression of disease signs. Microscopic examination of tissues collected at the time of euthanasia revealed massive accumulations of vacuolar inclusions in cells throughout the central nervous system, suggestive of a lysosomal storage disorder. A whole genome sequence generated with DNA from the affected dog contained a likely causal, homozygous missense variant in MAN2B1 that predicted an Asp104Gly amino acid substitution that was unique among whole genome sequences from over 4000 dogs. A lack of detectable α-mannosidase enzyme activity confirmed a diagnosis of a-mannosidosis. In addition to the vacuolar inclusions characteristic of α-mannosidosis, the dog exhibited accumulations of autofluorescent intracellular inclusions in some of the same tissues. The autofluorescence was similar to that which occurs in a group of lysosomal storage disorders called neuronal ceroid lipofuscinoses (NCLs). As in many of the NCLs, some of the storage bodies immunostained strongly for mitochondrial ATP synthase subunit c protein. This protein is not a substrate for α-mannosidase, so its accumulation and the development of storage body autofluorescence were likely due to a generalized impairment of lysosomal function secondary to the accumulation of α-mannosidase substrates. Thus, it appears that storage body autofluorescence and subunit c accumulation are not unique to the NCLs. Consistent with generalized lysosomal impairment, the affected dog exhibited accumulations of intracellular inclusions with varied and complex ultrastructural features characteristic of autophagolysosomes. Impaired autophagic flux may be a general feature of this class of disorders that contributes to disease pathology and could be a target for therapeutic intervention. In addition to storage body accumulation, glial activation indicative of neuroinflammation was observed in the brain and spinal cord of the proband.
Full article
(This article belongs to the Topic Animal Models of Human Disease)
►▼
Show Figures

Figure 1
Open AccessArticle
Diprosopus: A Rare Case of Craniofacial Duplication and a Systematic Review of the Literature
by
, , , , , , , , , and
Genes 2023, 14(9), 1745; https://doi.org/10.3390/genes14091745 - 31 Aug 2023
Abstract
In 1990, Gorlin et al. described four types of craniofacial duplications: (1) single mouth with duplication of the maxillary arch; (2) supernumerary mouth laterally placed with rudimentary segments; (3) single mouth with replication of the mandibular segments; and (4) true facial duplication, namely
[...] Read more.
In 1990, Gorlin et al. described four types of craniofacial duplications: (1) single mouth with duplication of the maxillary arch; (2) supernumerary mouth laterally placed with rudimentary segments; (3) single mouth with replication of the mandibular segments; and (4) true facial duplication, namely diprosopus. We describe a newborn born with wide-spaced eyes, a very broad nose, and two separate mouths. Workup revealed the absence of the corpus callosum and the presence of a brain midline lipoma, wide sutures, and a Chiari I malformation with cerebellar herniation. We conducted a systematic review of the literature and compared all the cases described as diprosopus. In 96% of these, the central nervous system is affected, with anencephaly being the most commonly associated abnormality. Other associated anomalies include cardiac malformations (86%), cleft palate (63%), diaphragmatic hernia (13%), and disorder of sex development (DSD) (13%). Although the facial features are those that first strike the eye, the almost obligate presence of cerebral malformations suggests a disruptive event in the cephalic pole of the forming embryo. No major monogenic contribution has been recognized today for this type of malformation.
Full article
(This article belongs to the Collection Study on Genotypes and Phenotypes of Pediatric Clinical Rare Diseases)
►▼
Show Figures
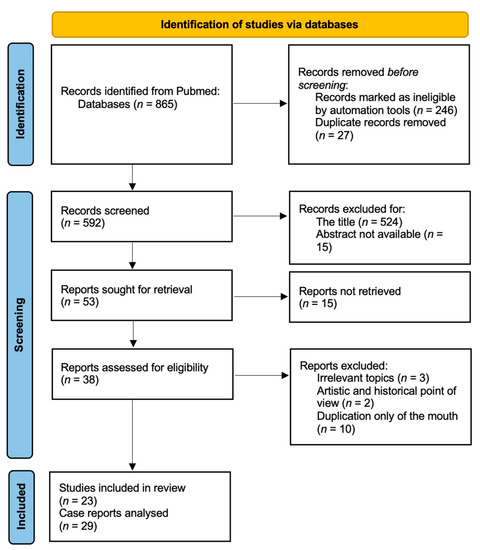
Figure 1
Open AccessCase Report
Demonstration of Parthenogenetic Reproduction in a Pet Ball Python (Python regius) through Analysis of Early-Stage Embryos
by
, , , , , , , and
Genes 2023, 14(9), 1744; https://doi.org/10.3390/genes14091744 - 31 Aug 2023
Abstract
Parthenogenesis is an asexual form of reproduction, normally present in various animal and plant species, in which an embryo is generated from a single gamete. Currently, there are some species for which parthenogenesis is supposed but not confirmed, and the mechanisms that activate
[...] Read more.
Parthenogenesis is an asexual form of reproduction, normally present in various animal and plant species, in which an embryo is generated from a single gamete. Currently, there are some species for which parthenogenesis is supposed but not confirmed, and the mechanisms that activate it are not well understood. A 10-year-old, wild-caught female ball python (Python regius) laid four eggs without any prior contact with a male. The eggs were not incubated and, after 3 days, were submitted to the University of Parma for analysis due to the suspicion of potential embryo presence. Examination of the egg content revealed residual blood vessels and a small red spot, indicative of an early-stage embryo. DNA was extracted from the three deceased embryos and from the mother’s blood, five microsatellites were analyzed to ascertain the origin of the embryos. The captive history data, together with the genetic microsatellite analysis approach, demonstrated the parthenogenetic origin of all three embryos. The embryos were homozygous for each of the maternal microsatellites, suggesting a terminal fusion automixis mode of development.
Full article
(This article belongs to the Special Issue Breeding and Functional Genomics in Animals)
Open AccessArticle
Comparative Plastomes of Curcuma alismatifolia (Zingiberaceae) Reveal Diversified Patterns among 56 Different Cut-Flower Cultivars
by
, , , , , , , , and
Genes 2023, 14(9), 1743; https://doi.org/10.3390/genes14091743 - 31 Aug 2023
Abstract
Curcuma alismatifolia (Zingiberaceae) is an ornamental species with high economic value due to its recent rise in popularity among floriculturists. Cultivars within this species have mixed genetic backgrounds from multiple hybridization events and can be difficult to distinguish via morphological and histological methods
[...] Read more.
Curcuma alismatifolia (Zingiberaceae) is an ornamental species with high economic value due to its recent rise in popularity among floriculturists. Cultivars within this species have mixed genetic backgrounds from multiple hybridization events and can be difficult to distinguish via morphological and histological methods alone. Given the need to improve identification resources, we carried out the first systematic study using plastomic data wherein genomic evolution and phylogenetic relationships from 56 accessions of C. alismatifolia were analyzed. The newly assembled plastomes were highly conserved and ranged from 162,139 bp to 164,111 bp, including 79 genes that code for proteins, 30 tRNA genes, and 4 rRNA genes. The A/T motif was the most common of SSRs in the assembled genomes. The Ka/Ks values of most genes were less than 1, and only two genes had Ka/Ks values above 1, which were rps15 (1.15), and ndhl (1.13) with petA equal to 1. The sequence divergence between different varieties of C. alismatifolia was large, and the percentage of variation in coding regions was lower than that in the non-coding regions. Such data will improve cultivar identification, marker assisted breeding, and preservation of germplasm resources.
Full article
(This article belongs to the Special Issue Advances in Evolution of Plant Organelle Genome (Volume II))
►▼
Show Figures
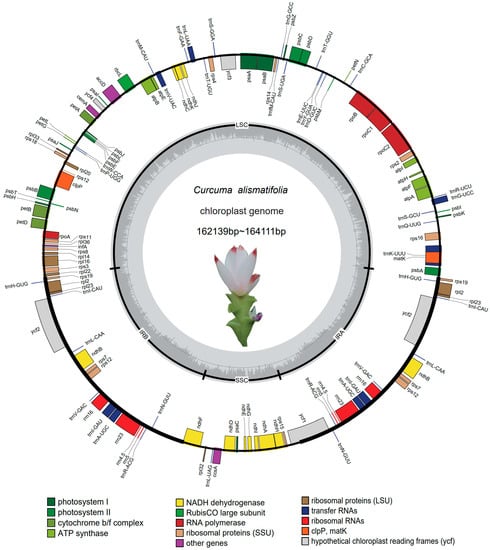
Figure 1
Open AccessArticle
An Automated Prognostic Model for Pancreatic Ductal Adenocarcinoma
by
, , , , , and
Genes 2023, 14(9), 1742; https://doi.org/10.3390/genes14091742 - 31 Aug 2023
Abstract
Pancreatic ductal adenocarcinoma (PDAC) constitutes a leading cause of cancer-related mortality despite advances in detection and treatment methods. While computed tomography (CT) serves as the current gold standard for initial evaluation of PDAC, its prognostic value remains limited, as it relies on diagnostic
[...] Read more.
Pancreatic ductal adenocarcinoma (PDAC) constitutes a leading cause of cancer-related mortality despite advances in detection and treatment methods. While computed tomography (CT) serves as the current gold standard for initial evaluation of PDAC, its prognostic value remains limited, as it relies on diagnostic stage parameters encompassing tumor size, lymph node involvement, and metastasis. Radiomics have recently shown promise in predicting postoperative survival of PDAC patients; however, they rely on manual pancreas and tumor delineation by clinicians. In this study, we collected a dataset of pre-operative CT scans from a cohort of 40 PDAC patients to evaluate a fully automated pipeline for survival prediction. Employing nnU-Net trained on an external dataset, we generated automated pancreas and tumor segmentations. Subsequently, we extracted 854 radiomic features from each segmentation, which we narrowed down to 29 via feature selection. We then combined these features with the Tumor, Node, Metastasis (TNM) system staging parameters, as well as the patient’s age. We trained a random survival forest model to perform an overall survival prediction over time, as well as a random forest classifier for the binary classification of two-year survival, using repeated cross-validation for evaluation. Our results exhibited promise, with a mean C-index of 0.731 for survival modeling and a mean accuracy of 0.76 in two-year survival prediction, providing evidence of the feasibility and potential efficacy of a fully automated pipeline for PDAC prognostication. By eliminating the labor-intensive manual segmentation process, our streamlined pipeline demonstrates an efficient and accurate prognostication process, laying the foundation for future research endeavors.
Full article
(This article belongs to the Special Issue Bioinformatics and Computational Biology for Cancer Prediction and Prognosis)
►▼
Show Figures
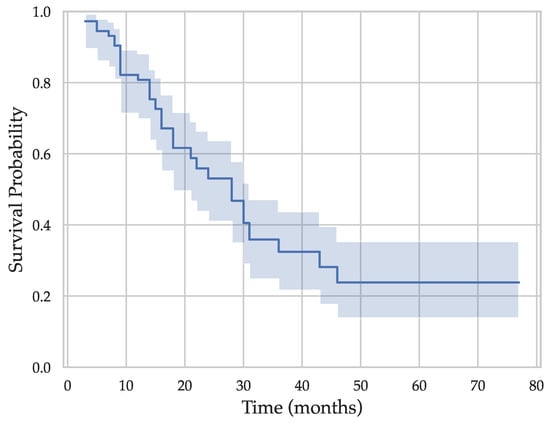
Figure 1
Open AccessReview
Nonribosomal Peptide Synthetases in Animals
by
, , , , and
Genes 2023, 14(9), 1741; https://doi.org/10.3390/genes14091741 - 30 Aug 2023
Abstract
Nonribosomal peptide synthetases (NRPSs) are a class of cytosolic enzymes that synthesize a range of bio-active secondary metabolites including antibiotics and siderophores. They are widespread among both prokaryotes and eukaryotes but are considered rare among animals. Recently, several novel NRPS genes have been
[...] Read more.
Nonribosomal peptide synthetases (NRPSs) are a class of cytosolic enzymes that synthesize a range of bio-active secondary metabolites including antibiotics and siderophores. They are widespread among both prokaryotes and eukaryotes but are considered rare among animals. Recently, several novel NRPS genes have been described in nematodes, schistosomes, and arthropods, which led us to investigate how prevalent NRPS genes are in the animal kingdom. We screened 1059 sequenced animal genomes and showed that NRPSs were present in 7 out of the 19 phyla analyzed. A phylogenetic analysis showed that the identified NRPSs form clades distinct from other adenylate-forming enzymes that contain similar domains such as fatty acid synthases. NRPSs show a remarkably scattered distribution over the animal kingdom. They are especially abundant in rotifers and nematodes. In rotifers, we found a large variety of domain architectures and predicted substrates. In the nematode Plectus sambesii, we identified the beta-lactam biosynthesis genes L-δ-(α-aminoadipoyl)-L-cysteinyl-D-valine synthetase, isopenicillin N synthase, and deacetoxycephalosporin C synthase that catalyze the formation of beta-lactam antibiotics in fungi and bacteria. These genes are also present in several species of Collembola, but not in other hexapods analyzed so far. In conclusion, our survey showed that NRPS genes are more abundant and widespread in animals than previously known.
Full article
(This article belongs to the Special Issue Review Collection on Population and Evolutionary Genetics and Genomics)
►▼
Show Figures
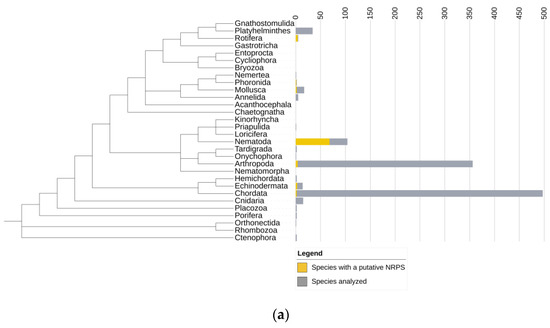
Figure 1
Open AccessArticle
Genetic Characterization of Blood Group Antigens for Polynesian Heritage Norfolk Island Residents
by
, , , , , , , , and
Genes 2023, 14(9), 1740; https://doi.org/10.3390/genes14091740 - 30 Aug 2023
Abstract
Improvements in blood group genotyping methods have allowed large scale population-based blood group genetics studies, facilitating the discovery of rare blood group antigens. Norfolk Island, an external and isolated territory of Australia, is one example of an underrepresented segment of the broader Australian
[...] Read more.
Improvements in blood group genotyping methods have allowed large scale population-based blood group genetics studies, facilitating the discovery of rare blood group antigens. Norfolk Island, an external and isolated territory of Australia, is one example of an underrepresented segment of the broader Australian population. Our study utilized whole genome sequencing data to characterize 43 blood group systems in 108 Norfolk Island residents. Blood group genotypes and phenotypes across the 43 systems were predicted using RBCeq. Predicted frequencies were compared to data available from the 1000G project. Additional copy number variation analysis was performed, investigating deletions outside of RHCE, RHD, and MNS systems. Examination of the ABO blood group system predicted a higher distribution of group A1 (45.37%) compared to group O (35.19%) in residents of the Norfolk Island group, similar to the distribution within European populations (42.94% and 38.97%, respectively). Examination of the Kidd blood group system demonstrated an increased prevalence of variants encoding the weakened Kidd phenotype at a combined prevalence of 12.04%, which is higher than that of the European population (5.96%) but lower than other populations in 1000G. Copy number variation analysis showed deletions within the Chido/Rodgers and ABO blood group systems. This study is the first step towards understanding blood group genotype and antigen distribution on Norfolk Island.
Full article
(This article belongs to the Special Issue Population and Diseases Genomics)
►▼
Show Figures
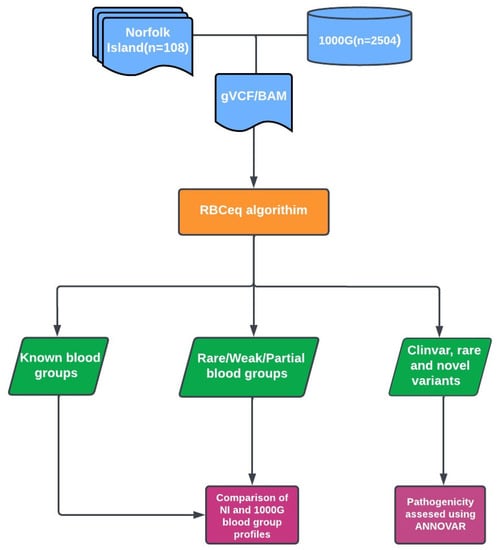
Figure 1
Open AccessArticle
Screening of Reference Genes under Biotic Stress and Hormone Treatment of Mung Bean (Vigna radiata) by Quantitative Real-Time PCR
by
, , , , , , , and
Genes 2023, 14(9), 1739; https://doi.org/10.3390/genes14091739 - 30 Aug 2023
Abstract
Mung bean (Vigna radiata) production has been greatly threatened by numerous diseases. Infection with these pathogens causes extensive changes in gene expression and the activation of hormone signal transduction. Quantitative real-time PCR (qRT-PCR) is the most common technique used for gene
[...] Read more.
Mung bean (Vigna radiata) production has been greatly threatened by numerous diseases. Infection with these pathogens causes extensive changes in gene expression and the activation of hormone signal transduction. Quantitative real-time PCR (qRT-PCR) is the most common technique used for gene expression validation. Screening proper reference genes for mung bean under pathogen infection and hormone treatment is a prerequisite for ensuring the accuracy of qRT-PCR data in mung bean disease-resistance research. In this study, six candidate reference genes (Cons4, ACT, TUA, TUB, GAPDH, and EF1α) were selected to evaluate the expression stability under four soil-borne disease pathogens (Pythium myriotylum, Pythium aphanidermatum, Fusarium oxysporum, and Rhizoctonia solani) and five hormone treatments (SA, MeJA, ETH, ABA, and GA3). In the samples from different treatments, the Ct value distribution of the six candidate reference genes was different. Under the condition of hormone treatment, the Ct value ranged from a minimum of 17.87 for EF1α to a maximum of 29.63 for GAPDH. Under the condition of pathogen infection, the Ct value ranged from a minimum of 19.43 for EF1α to a maximum of 31.82 for GAPDH. After primer specificity analysis, it was found that GAPDH was not specific, so the five reference genes Cons4, ACT, TUA, TUB, and EF1α were used in subsequent experiments. The software products GeNorm, NormFinder, BestKeeper and RefFinder were used for qRT-PCR data analysis. In general, the best candidates reference genes were: TUA for SA, ABA, GA3, and Pythium myriotylum treatment; TUB for ETH treatment; ACT for MeJA and Fusarium oxysporum treatment; and EF1α for Pythium aphanidermatum and Rhizoctonia solani treatment. The most stably expressed genes in all samples were TUA, while Cons4 was the least stable reference gene. Finally, the reliability of the reference gene was further validated by analysis of the expression profiles of four mung bean genes (Vradi0146s00260, Vradi0158s00480, Vradi07g23860, and Vradi11g03350) selected from transcriptome data. Our results provide more accurate information for the normalization of qRT-PCR data in mung bean response to pathogen interaction.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
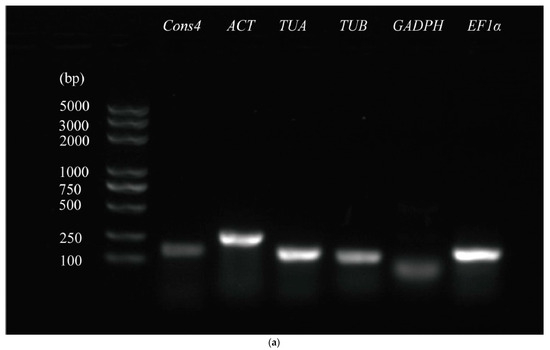
Figure 1
Open AccessArticle
Comparative Mitogenomic Analyses of Darkling Beetles (Coleoptera: Tenebrionidae) Provide Evolutionary Insights into tRNA-like Sequences
by
, , , , , , and
Genes 2023, 14(9), 1738; https://doi.org/10.3390/genes14091738 - 30 Aug 2023
Abstract
Tenebrionidae is widely recognized owing to its species diversity and economic importance. Here, we determined the mitochondrial genomes (mitogenomes) of three Tenebrionidae species (Melanesthes exilidentata, Anatolica potanini, and Myladina unguiculina) and performed a comparative mitogenomic analysis to characterize the
[...] Read more.
Tenebrionidae is widely recognized owing to its species diversity and economic importance. Here, we determined the mitochondrial genomes (mitogenomes) of three Tenebrionidae species (Melanesthes exilidentata, Anatolica potanini, and Myladina unguiculina) and performed a comparative mitogenomic analysis to characterize the evolutionary characteristics of the family. The tenebrionid mitogenomes were highly conserved with respect to genome size, gene arrangement, base composition, and codon usage. All protein-coding genes evolved under purifying selection. The largest non-coding region (i.e., control region) showed several unusual features, including several conserved repetitive fragments (e.g., A+T-rich regions, G+C-rich regions, Poly-T tracts, TATA repeat units, and longer repetitive fragments) and tRNA-like structures. These tRNA-like structures can bind to the appropriate anticodon to form a cloverleaf structure, although base-pairing is not complete. We summarized the quantity, types, and conservation of tRNA-like sequences and performed functional and evolutionary analyses of tRNA-like sequences with various anticodons. Phylogenetic analyses based on three mitogenomic datasets and two tree inference methods largely supported the monophyly of each of the three subfamilies (Stenochiinae, Pimeliinae, and Lagriinae), whereas both Tenebrioninae and Diaperinae were consistently recovered as polyphyletic. We obtained a tenebrionid mitogenomic phylogeny: (Lagriinae, (Pimeliinae, ((Tenebrioninae + Diaperinae), Stenochiinae))). Our results provide insights into the evolution and function of tRNA-like sequences in tenebrionid mitogenomes and contribute to our general understanding of the evolution of Tenebrionidae.
Full article
(This article belongs to the Special Issue Genetics, Phylogeny, and Evolution of Insects)
►▼
Show Figures
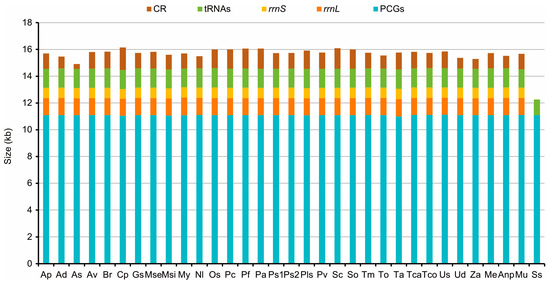
Figure 1
Open AccessArticle
Characterization of Plant Homeodomain Transcription Factor Genes Involved in Flower Development and Multiple Abiotic Stress Response in Pepper
by
, , , , , , , , and
Genes 2023, 14(9), 1737; https://doi.org/10.3390/genes14091737 (registering DOI) - 30 Aug 2023
Abstract
Plant homeodomain (PHD) transcription factor genes are involved in plant development and in a plant’s response to stress. However, there are few reports about this gene family in peppers (Capsicum annuum L.). In this study, the pepper inbred line “Zunla-1” was used
[...] Read more.
Plant homeodomain (PHD) transcription factor genes are involved in plant development and in a plant’s response to stress. However, there are few reports about this gene family in peppers (Capsicum annuum L.). In this study, the pepper inbred line “Zunla-1” was used as the reference genome, and a total of 43 PHD genes were identified, and systematic analysis was performed to study the chromosomal location, evolutionary relationship, gene structure, domains, and upstream cis-regulatory elements of the CaPHD genes. The fewest CaPHD genes were located on chromosome 4, while the most were on chromosome 3. Genes with similar gene structures and domains were clustered together. Expression analysis showed that the expression of CaPHD genes was quite different in different tissues and in response to various stress treatments. The expression of CaPHD17 was different in the early stage of flower bud development in the near-isogenic cytoplasmic male-sterile inbred and the maintainer inbred lines. It is speculated that this gene is involved in the development of male sterility in pepper. CaPHD37 was significantly upregulated in leaves and roots after heat stress, and it is speculated that CaPHD37 plays an important role in tolerating heat stress in pepper; in addition, CaPHD9, CaPHD10, CaPHD11, CaPHD17, CaPHD19, CaPHD20, and CaPHD43 were not sensitive to abiotic stress or hormonal factors. This study will provide the basis for further research into the function of CaPHD genes in plant development and responses to abiotic stresses and hormones.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
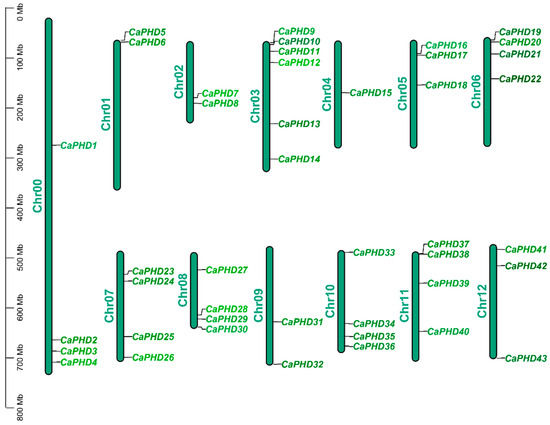
Figure 1
Open AccessReview
Non-Coding RNAs and Gut Microbiota in the Pathogenesis of Cardiac Arrhythmias: The Latest Update
Genes 2023, 14(9), 1736; https://doi.org/10.3390/genes14091736 - 30 Aug 2023
Abstract
Non-coding RNAs (ncRNAs) are indispensable for adjusting gene expression and genetic programming throughout development and for health as well as cardiovascular diseases. Cardiac arrhythmia is a frequent cardiovascular disease that has a complex pathology. Recent studies have shown that ncRNAs are also associated
[...] Read more.
Non-coding RNAs (ncRNAs) are indispensable for adjusting gene expression and genetic programming throughout development and for health as well as cardiovascular diseases. Cardiac arrhythmia is a frequent cardiovascular disease that has a complex pathology. Recent studies have shown that ncRNAs are also associated with cardiac arrhythmias. Many non-coding RNAs and/or genomes have been reported as genetic background for cardiac arrhythmias. In general, arrhythmias may be affected by several functional and structural changes in the myocardium of the heart. Therefore, ncRNAs might be indispensable regulators of gene expression in cardiomyocytes, which could play a dynamic role in regulating the stability of cardiac conduction and/or in the remodeling process. Although it remains almost unclear how ncRNAs regulate the expression of molecules for controlling cardiac conduction and/or the remodeling process, the gut microbiota and immune system within the intricate networks might be involved in the regulatory mechanisms. This study would discuss them and provide a research basis for ncRNA modulation, which might support the development of emerging innovative therapies against cardiac arrhythmias.
Full article
(This article belongs to the Special Issue Genetics of Human Cardiovascular Disease)
►▼
Show Figures
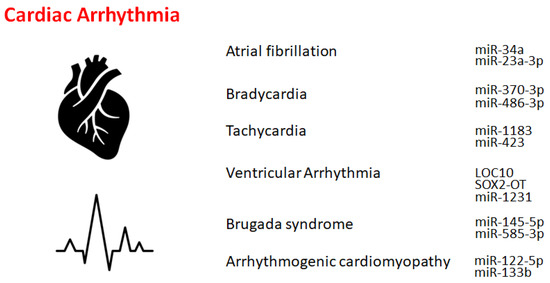
Figure 1
Open AccessReview
The Stem Cell Expression Profile of Odontogenic Tumors and Cysts: A Systematic Review and Meta-Analysis
by
, , , , , and
Genes 2023, 14(9), 1735; https://doi.org/10.3390/genes14091735 - 30 Aug 2023
Abstract
Background: Stem cells have been associated with self-renewing and plasticity and have been investigated in various odontogenic lesions in association with their pathogenesis and biological behavior. We aim to provide a systematic review of stem cell markers’ expression in odontogenic tumors and cysts.
[...] Read more.
Background: Stem cells have been associated with self-renewing and plasticity and have been investigated in various odontogenic lesions in association with their pathogenesis and biological behavior. We aim to provide a systematic review of stem cell markers’ expression in odontogenic tumors and cysts. Methods: The literature was searched through the MEDLINE/PubMed, EMBASE via OVID, Web of Science, and CINHAL via EBSCO databases for original studies evaluating stem cell markers’ expression in different odontogenic tumors/cysts, or an odontogenic disease group and a control group. The studies’ risk of bias (RoB) was assessed via a Joanna Briggs Institute Critical Appraisal Tool. Meta-analysis was conducted for markers evaluated in the same pair of odontogenic tumors/cysts in at least two studies. Results: 29 studies reported the expression of stem cell markers, e.g., SOX2, OCT4, NANOG, CD44, ALDH1, BMI1, and CD105, in various odontogenic lesions, through immunohistochemistry/immunofluorescence, polymerase chain reaction, flow cytometry, microarrays, and RNA-sequencing. Low, moderate, and high RoBs were observed in seven, nine, and thirteen studies, respectively. Meta-analysis revealed a remarkable discriminative ability of SOX2 for ameloblastic carcinomas or odontogenic keratocysts over ameloblastomas. Conclusion: Stem cells might be linked to the pathogenesis and clinical behavior of odontogenic pathologies and represent a potential target for future individualized therapies.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
►▼
Show Figures
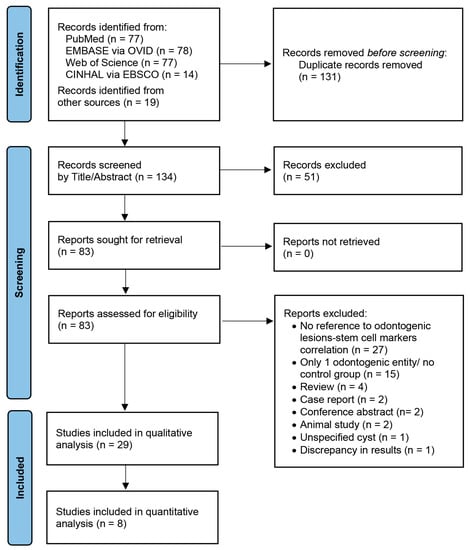
Figure 1
Open AccessReview
Conceptualizing Epigenetics and the Environmental Landscape of Autism Spectrum Disorders
by
, , , , , , , , , and
Genes 2023, 14(9), 1734; https://doi.org/10.3390/genes14091734 - 30 Aug 2023
Abstract
Complex interactions between gene variants and environmental risk factors underlie the pathophysiological pathways in major psychiatric disorders. Autism Spectrum Disorder is a neuropsychiatric condition in which susceptible alleles along with epigenetic states contribute to the mutational landscape of the ailing brain. The present
[...] Read more.
Complex interactions between gene variants and environmental risk factors underlie the pathophysiological pathways in major psychiatric disorders. Autism Spectrum Disorder is a neuropsychiatric condition in which susceptible alleles along with epigenetic states contribute to the mutational landscape of the ailing brain. The present work reviews recent evolutionary, molecular, and epigenetic mechanisms potentially linked to the etiology of autism. First, we present a clinical vignette to describe clusters of maladaptive behaviors frequently diagnosed in autistic patients. Next, we microdissect brain regions pertinent to the nosology of autism, as well as cell networks from the bilateral body plan. Lastly, we catalog a number of pathogenic environments associated with disease risk factors. This set of perspectives provides emerging insights into the dynamic interplay between epigenetic and environmental variation in the development of Autism Spectrum Disorders.
Full article
(This article belongs to the Special Issue Genes, Phenotypes and Molecular Mechanisms for Personalized Medicine in Autism)
►▼
Show Figures
Figure 1
Open AccessArticle
Mechanisms of Regulation of the CHRDL1 Gene by the TWIST2 and ADD1/SREBP1c Transcription Factors
Genes 2023, 14(9), 1733; https://doi.org/10.3390/genes14091733 - 30 Aug 2023
Abstract
Setleis syndrome (SS) is a rare focal facial dermal dysplasia caused by recessive mutations in the basic helix-loop-helix (bHLH) transcription factor, TWIST2. Expression microarray analysis showed that the chordin-like 1 (CHRDL1) gene is up-regulated in dermal fibroblasts from three SS patients
[...] Read more.
Setleis syndrome (SS) is a rare focal facial dermal dysplasia caused by recessive mutations in the basic helix-loop-helix (bHLH) transcription factor, TWIST2. Expression microarray analysis showed that the chordin-like 1 (CHRDL1) gene is up-regulated in dermal fibroblasts from three SS patients with the Q119X TWIST2 mutation. METHODS: Putative TWIST binding sites were found in the upstream region of the CHRDL1 gene and examined by electrophoretic mobility shift (EMSA) and reporter gene assays. RESULTS: EMSAs showed specific binding of TWIST1 and TWIST2 homodimers, as well as heterodimers with E12, to the more distal E-boxes. An adjoining E-box was bound by ADD1/SREBP1c. EMSA analysis suggested that TWIST2 and ADD1/SREBP1c could compete for binding. Luciferase (luc) reporter assays revealed that the CHRDL1 gene upstream region drives its expression and ADD1/SREBP1c increased it 2.6 times over basal levels. TWIST2, but not the TWIST2-Q119X mutant, blocked activation by ADD1/SREBP1c, but overexpression of TWIST2-Q119X increased luc gene expression. In addition, EMSA competition assays showed that TWIST2, but not TWIST1, competes with ADD1/SREBP1c for DNA binding to the same site. CONCLUSIONS: Formation of an inactive complex between the TWIST2 Q119X and Q65X mutant proteins and ADD1/SREBP1c may prevent repressor binding and allow the binding of other regulators to activate CHRDL1 gene expression.
Full article
(This article belongs to the Section Human Genomics and Genetic Diseases)
►▼
Show Figures
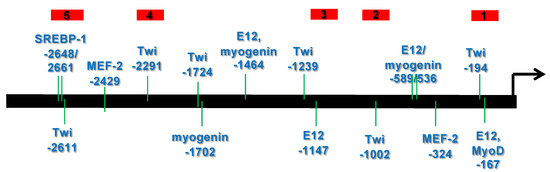
Figure 1
Open AccessArticle
Maize OPR2 and LOX10 Mediate Defense against Fall Armyworm and Western Corn Rootworm by Tissue-Specific Regulation of Jasmonic Acid and Ketol Metabolism
by
, , , , , and
Genes 2023, 14(9), 1732; https://doi.org/10.3390/genes14091732 - 30 Aug 2023
Abstract
Foliage-feeding fall armyworm (FAW; Spodoptera frugiperda) and root-feeding western corn rootworm (WCR; Diabrotica virgifera virgifera) are maize (Zea mays L.) pests that cause significant yield losses. Jasmonic acid (JA) plays a pivotal defense role against insects. 12-oxo-phytodienoic acid (12-OPDA) is
[...] Read more.
Foliage-feeding fall armyworm (FAW; Spodoptera frugiperda) and root-feeding western corn rootworm (WCR; Diabrotica virgifera virgifera) are maize (Zea mays L.) pests that cause significant yield losses. Jasmonic acid (JA) plays a pivotal defense role against insects. 12-oxo-phytodienoic acid (12-OPDA) is converted into JA by peroxisome-localized OPDA reductases (OPR). However, little is known about the physiological functions of cytoplasmic OPRs. Here, we show that disruption of ZmOPR2 reduced wound-induced JA production and defense against FAW while accumulating more JA catabolites. Overexpression of ZmOPR2 in Arabidopsis enhanced JA production and defense against beet armyworm (BAW; Spodoptera exigua). In addition, lox10opr2 double mutants were more susceptible than either single mutant, suggesting that ZmOPR2 and ZmLOX10 uniquely and additively contributed to defense. In contrast to the defensive roles of ZmOPR2 and ZmLOX10 in leaves, single mutants did not display any alteration in root herbivory defense against WCR. Feeding on lox10opr2 double mutants resulted in increased WCR mortality associated with greater herbivory-induced production of insecticidal death acids and ketols. Thus, ZmOPR2 and ZmLOX10 cooperatively inhibit the synthesis of these metabolites during herbivory by WCR. We conclude that ZmOPR2 and ZmLOX10 regulate JA-mediated resistance in leaves against FAW while suppressing insecticidal oxylipin synthesis in roots during WCR infestation.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
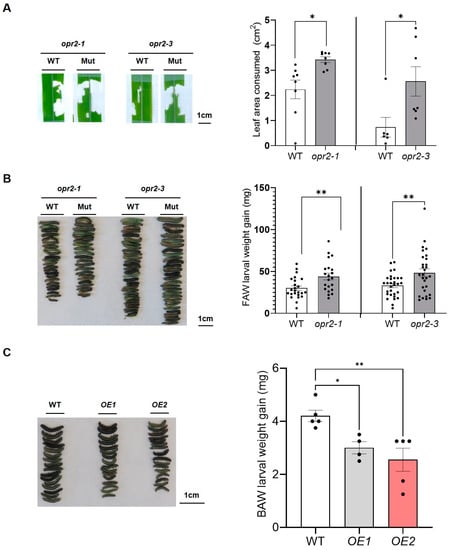
Figure 1
Open AccessArticle
Genomic Characterization of a Tetracycline-Resistant Strain of Brochothrix thermosphacta Highlights Plasmids Partially Shared between Various Strains
Genes 2023, 14(9), 1731; https://doi.org/10.3390/genes14091731 - 30 Aug 2023
Abstract
The Gram-positive bacterium Brochothrix thermosphacta is a spoilage agent commonly found on meat products. While the tet(L) gene, which confers resistance to tetracycline, has been identified in certain strains of B. thermosphacta, only a limited number of studies have investigated this
[...] Read more.
The Gram-positive bacterium Brochothrix thermosphacta is a spoilage agent commonly found on meat products. While the tet(L) gene, which confers resistance to tetracycline, has been identified in certain strains of B. thermosphacta, only a limited number of studies have investigated this gene and its potential presence on mobile DNA elements. This study aims to analyze the tetracycline-resistant strain B. thermosphacta BT469 at the genomic level to gain insight into the molecular determinants responsible for this resistance. Three plasmids have been identified in the strain: pBT469-1, which contains a tetR gene; pBT469-2, which harbours the tet(L) gene responsible for tetracycline resistance; and pBT469-3, which carries genes encoding for a thioredoxin and a phospholipase A2. Homology searches among sequences in public databases have revealed that the plasmid pBT469-2 is currently unique to the BT469 strain. However, the pBT469-1 plasmid is also found in three other strains of B. thermosphacta. Notably, sequences similar to pBT469-1 and pBT469-2 were also found in other bacterial genera, suggesting that these plasmids may be part of a diverse family present in several bacterial genera. Interestingly, sequences of various strains of B. thermosphacta show a high level of similarity with pBT469-3, suggesting that variants of this plasmid could be frequently found in this bacterium.
Full article
(This article belongs to the Section Microbial Genetics and Genomics)
►▼
Show Figures
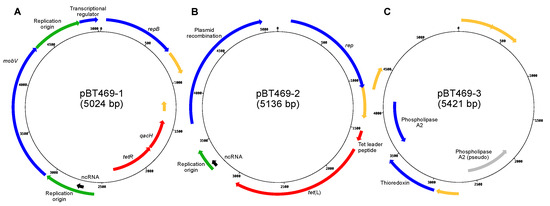
Figure 1
Open AccessBrief Report
Thymoquinone Potentially Modulates the Expression of Key Onco- and Tumor Suppressor miRNAs in Prostate and Colon Cancer Cell Lines: Insights from PC3 and HCT-15 Cells
by
, , , , , and
Genes 2023, 14(9), 1730; https://doi.org/10.3390/genes14091730 - 30 Aug 2023
Abstract
Prostate cancer (PC) and colon cancer significantly contribute to global cancer-related morbidity and mortality. Thymoquinone (TQ), a naturally occurring phytochemical found in black cumin, has shown potential as an anticancer compound. This study aimed to investigate the effects of TQ on the expression
[...] Read more.
Prostate cancer (PC) and colon cancer significantly contribute to global cancer-related morbidity and mortality. Thymoquinone (TQ), a naturally occurring phytochemical found in black cumin, has shown potential as an anticancer compound. This study aimed to investigate the effects of TQ on the expression profile of key tumor suppressor and onco-suppressor miRNAs in PC3 prostate cancer cells and HCT-15 colon cancer cells. Cell viability assays revealed that TQ inhibited the growth of both cell lines in a dose-dependent manner, with IC50 values of approximately 82.59 μM for HCT-15 and 55.83 μM for PC3 cells. Following TQ treatment at the IC50 concentrations, miRNA expression analysis demonstrated that TQ significantly downregulated miR-21-5p expression in HCT-15 cells and upregulated miR-34a-5p, miR-221-5p, miR-17-5p, and miR-21-5p expression in PC3 cells. However, no significant changes were observed in the expression levels of miR-34a-5p and miR-200a-5p in HCT-15 cells. The current findings suggest that TQ might exert its antiproliferative effects by modulating specific tumor suppressor and onco-suppressor miRNAs in prostate and colon cancer cells. Further investigations are warranted to elucidate the precise underlying mechanisms and to explore the therapeutic potential of TQ in cancer treatment. To the best of our knowledge, this is the first report regarding the effect of TQ on the miRNA expression profile in colon and prostate cancer cell lines.
Full article
(This article belongs to the Special Issue Non-coding RNAs in Human Health and Disease)
►▼
Show Figures
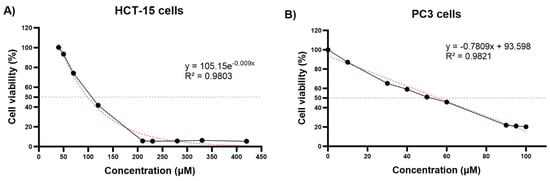
Figure 1

Journal Menu
► ▼ Journal Menu-
- Genes Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Cells, Epigenomes, Genes, IJMS, IJTM
Stem Cell Differentiation and Applications
Topic Editors: Hiroyuki Hirai, Haiyun Pei, Atsushi AsakuraDeadline: 20 October 2023
Topic in
Applied Sciences, BioMedInformatics, BioTech, Genes, Computation
Computational Intelligence and Bioinformatics (CIB)
Topic Editors: Marco Mesiti, Giorgio Valentini, Elena Casiraghi, Tiffany J. CallahanDeadline: 31 October 2023
Topic in
AI, Entropy, Genes, IJMS, MAKE
Bioinformatics and Intelligent Information Processing
Topic Editors: Zhiping Liu, Han Zhang, Junwei HanDeadline: 30 November 2023
Topic in
Agriculture, Agronomy, Crops, Genes, Plants
Vegetable Breeding, Genetics and Genomics
Topic Editors: Umesh K. Reddy, Padma Nimmakayala, Georgia NtatsiDeadline: 31 December 2023

Conferences
Special Issues
Special Issue in
Genes
Pharmacogenomics: Challenges and Future
Guest Editors: Mariamena Arbitrio, Maria Teresa Di Martino, Francesca SciontiDeadline: 5 September 2023
Special Issue in
Genes
Systematic Analysis and Application of Omics Data in Animal Breeding
Guest Editors: Fuping Zhao, Zhe Zhang, Lingzhao Fang, Ruidong XiangDeadline: 15 September 2023
Special Issue in
Genes
Nutrigenomics and Cellular Metabolism
Guest Editor: Mario G. MirisolaDeadline: 1 October 2023
Special Issue in
Genes
Genetics and Genomics of Inherited Metabolic Diseases
Guest Editors: Ewa Piotrowska, Magdalena PodlachaDeadline: 15 October 2023
Topical Collections
Topical Collection in
Genes
Study on Genotypes and Phenotypes of Pediatric Clinical Rare Diseases
Collection Editors: Livia Garavelli, Stefano Giuseppe Caraffi
Topical Collection in
Genes
Eukaryotic Non-coding RNAs: Diversity, Structure/Function, Implication in Cardiovascular Disease
Collection Editors: Morten Andre Høydal, Christiane Branlant
Topical Collection in
Genes
Feature Papers in Animal Genetics and Genomics
Collection Editors: Antonio Figueras, Raquel Vasconcelos, Brenda Oppert
















